What is ATAC-Seq?
The assay for transposase-accessible chromatin with sequencing (ATAC-Seq) is a popular method for determining chromatin accessibility across the genome. By sequencing regions of open chromatin, ATAC-Seq can help you uncover how chromatin packaging and other factors affect gene expression.
ATAC-Seq does not require prior knowledge of regulatory elements, making it a powerful epigenetic discovery tool. It has been used to better understand chromatin accessibility, transcription factor binding, and gene regulation in complex diseases, embryonic development, T-cell activation, and cancer.1,2 ATAC-Seq can be performed on bulk cell populations or on single cells at high resolution.
Applications of ATAC-Seq
Chromatin accessibility analysis with ATAC-Seq can provide valuable insights into the regulatory landscape of the genome. Popular applications include:
- Nucleosome mapping
- Transcription factor binding analysis
- Novel enhancer identification
- Exploration of disease-relevant regulatory mechanisms
- Cell type–specific regulation analysis
- Evolutionary studies
- Comparative epigenomics
- Biomarker discovery
Additionally, ATAC-Seq can be combined with other methods, such as RNA sequencing, for a multiomic approach to studying gene expression.3 Subsequent experiments often include ChIP-Seq, Methyl-Seq, or Hi-C-Seq to further characterize forms of epigenetic regulation.
"ATAC-Seq allows you to ask questions about the epigenetic variability in complex or rare tissues and epigenomic landscape in populations of cells that haven’t been observable at the genome-wide level before."
How Does ATAC-Seq Work?
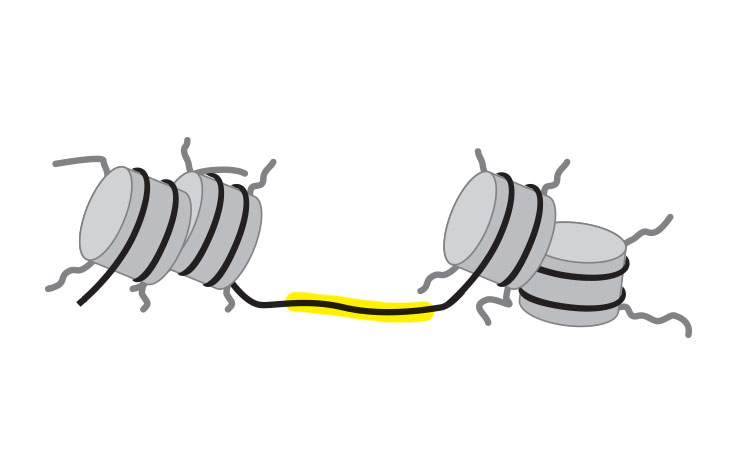
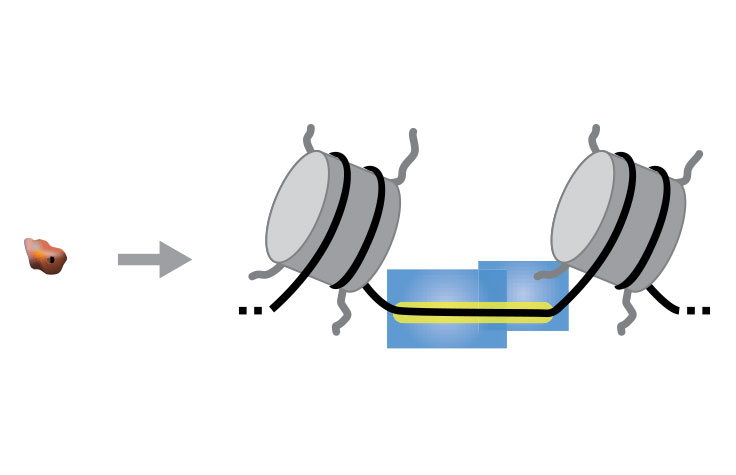
In ATAC-Seq, genomic DNA is exposed to Tn5, a highly active transposase. Tn5 simultaneously fragments DNA, preferentially inserts into open chromatin sites, and adds sequencing primers (a process known as tagmentation). The sequenced DNA identifies the open chromatin and data analysis can provide insight into gene regulation.

Surveying the Chromatin Landscape with ATAC-Seq
Learn how Dr. Greenleaf and his team developed ATAC-Seq and why he believes that it might one day provide new insights into the development and treatment of cancer and autoimmune disease.
Read InterviewATAC-Seq Protocol
We recommend the following ATAC-Seq protocol: Buenrostro J, Wu B, Chang H, Greenleaf W. ATAC-seq: a method for assaying chromatin accessibility genome-wide. Curr Protoc Mol Biol. 2015;109:21.29.1-21.29-9.
Note that the TDE1 Enzyme and Buffer Kits are now available separately from Illumina (Cat. No. 20034197 and 20034198). All other steps in the protocol, including the enzyme and buffer concentration, remain the same.
View Protocol
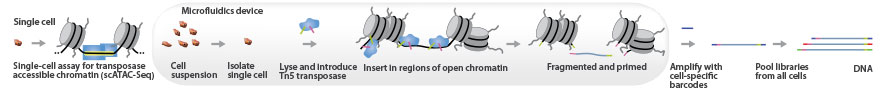
Single-Cell ATAC-Seq
Single-cell ATAC-Seq combines compartmentalization and barcoding of single cells with Tn5 tagmentation. The Tn5 transposase tags open chromatin regions with sequencing adapters. The tagged DNA fragments are then purified, amplified, and sequenced.
Learn more about single-cell sequencingCoverage Recommendations for ATAC-Seq
The minimum required sequencing coverage for ATAC-Seq varies according to research objectives. This table provides some guidelines for common applications.
We recommend using paired-end reads for ATAC-Seq. Compared to single-read sequencing, paired-end reads offer:
- Higher unique alignment rates
- Removal of PCR duplicates
- More complete information about accessible sequences
- Ability to categorize reads as nucleosome-free, mono-nucleosomal, or di-nucleosomal
| Research Goal | Recommended Depth |
|---|---|
| Identification of open chromatin differences in human samples | ≥ 50 M paired-end reads |
| Transcription factor foot printing to construct gene regulatory network | > 200 M paired-end reads |
| Single-cell analysis | 25–50 K paired-end reads per nucleus/cell |
As experimental needs can vary, we encourage you to consult the scientific literature to determine the right level of coverage for your project.
Webinar: Assaying Genome-Wide Chromatin Accessibility with ATAC-Seq
In this webinar, you'll learn how to use ATAC-seq for genome-wide chromatin accessibility profiling and how it fits in with other chromatin accessibility profiling methods. William Greenleaf discusses the development of ATAC-seq, its use, and adaptation to single cells. Bing Ren and Sebastian Preissl discuss single-cell ATAC-seq using combinatorial indexing, and application to brain, hearts and other tissues.
View WebinarFeatured ATAC-Seq Publications
Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position.
Buenrostro JD, Giresi PG, Zaba LC, et al.
Nat Methods. 2013;10(12):1213-1218.
Epigenome-wide study uncovers large-scale changes in histone acetylation driven by tau pathology in aging and Alzheimer’s human brains.
Klein HU, McCabe C, Gjoneska E, et al.
Nat Neurosci. 2019;22(1):37-46.
Epigenetic control of innate and adaptive immune memory.
Lau CM, Adams NM, Geary CD, et al.
Nat Immunol. 2018;19(9):963-972.
Multiplex single-cell profiling of chromatin accessibility by combinatorial cellular indexing.
Cusanovich DA, Daza R, Ade A, et al.
Science. 2015;348(6237):910-914.
Single-cell chromatin accessibility reveals principles of regulatory variation.
Buenrostro JD, Wu B, Litzenburger UM, et al.
Nature. 2015;523(7561):486-490.
Droplet-based combinatorial indexing for massive-scale single-cell chromatin accessibility.
Lareau CA, Duarte FM, Chew JG, et al.
Nat Biotechnol. 2019;37(8):916-924.
Featured Products
ATAC-Seq Workflow
Interested in receiving newsletters, case studies, and information on genomic analysis techniques? Enter your email address.
Related Methods & Applications
References
- Cusanovich DA, Reddington JP, Garfield DA, et al. The cis-regulatory dynamics of embryonic development at single-cell resolution. Nature. 2018; 555:538-542. 13.
- Gate RE, Cheng CS, Aiden AP, et al. Genetic determinants of co-accessible chromatin regions in activated T-cells across humans. Nat Genet. 2018; 50:1140-1150.
- Cao J, Cusanovich DA, Ramani V, et al. Joint profiling of chromatin accessibility and gene expression in thousands of single cells. Science. 2018;361:1380-1385.